library(palmerpenguins)
library(tidymodels)
library(tidyverse)
library(ggplot2)When learning things I always like to use Palmer Penguins! It’s a fun dataset and after doing so many projects with iris data or car data it’s nice to change it up.
Exploring Data
This is a data set of observations of penguins, and here is a tibble so we can get an overview of what we know about them.
penguins %>%
head()# A tibble: 6 × 7
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
# ℹ 1 more variable: sex <fct>I want to see what the counts for some things are to get a feel for how many NA’s I have as the tibble above shows that some entries are NA.
penguins %>%
count(species)# A tibble: 3 × 2
species n
<fct> <int>
1 Adelie 152
2 Chinstrap 68
3 Gentoo 124penguins %>%
count(sex)# A tibble: 3 × 2
sex n
<fct> <int>
1 female 165
2 male 168
3 <NA> 11penguins %>%
count(island, species)# A tibble: 5 × 3
island species n
<fct> <fct> <int>
1 Biscoe Adelie 44
2 Biscoe Gentoo 124
3 Dream Adelie 56
4 Dream Chinstrap 68
5 Torgersen Adelie 52Okay so I have ~11 entries with NA. I think for this model I’ll try to make a classification model to see if I can corrently predict the sex based on it’s characteristics. I am going to do a little more exploration to see how feasible this is.
penguins %>%
filter(!is.na(sex)) %>%
ggplot(aes(flipper_length_mm, bill_length_mm, color = sex, size = body_mass_g)) +
geom_point(alpha = .7) +
facet_wrap(~species)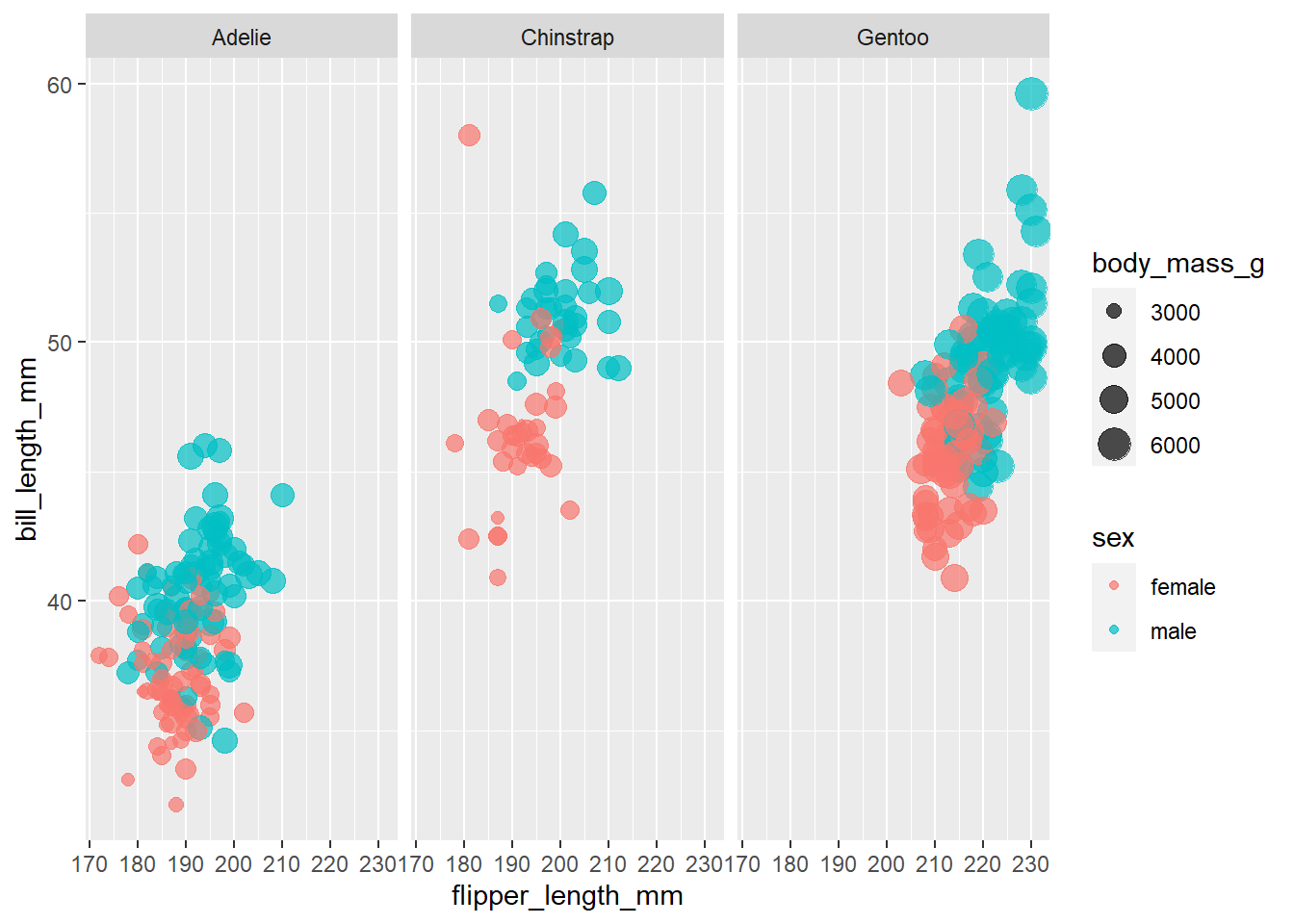
So notice that for all 3 species of penguins, female penguins have smaller bill lengths, flipper lengths and generally less body mass. There is some overlap, but it’s nothing very messy. This seems like some good indication that for all 3 species we can use these two feature to build our classifier. Also interesting that the smallest Gentoo is about as large as the biggest Adelie.
Building A Model
Let’s grab our data and store it in a data frame so we can start to build a model.
penguins_df <- penguins %>%
filter(!is.na(sex)) %>%
select(-island)Now to split the data into testing and training sets.
set.seed(777)
penguin_split <- initial_split(penguins_df, strata = sex)
penguin_training <- training(penguin_split)
penguin_testing <- testing(penguin_split)The default for initial split is a 75% split, and while this isn’t a large data set we can create a resampled data set using the training set to compute the performance of the models that I train. So using boostraps I will create a set of objects each with ~250 training samples and ~90 testing samples (all with resampling) to have a larget dataset to train my classifier with.
set.seed(2134)
penguin_boot <- bootstraps(penguin_training)
penguin_boot# Bootstrap sampling
# A tibble: 25 × 2
splits id
<list> <chr>
1 <split [249/91]> Bootstrap01
2 <split [249/89]> Bootstrap02
3 <split [249/89]> Bootstrap03
4 <split [249/84]> Bootstrap04
5 <split [249/93]> Bootstrap05
6 <split [249/82]> Bootstrap06
7 <split [249/89]> Bootstrap07
8 <split [249/94]> Bootstrap08
9 <split [249/92]> Bootstrap09
10 <split [249/96]> Bootstrap10
# ℹ 15 more rowsNow to make a basic logistic regression model and another model. What I am doing with this command is setting up what kind of model I want to train. I haven’t trained it, yet… the other will be a random forest because they usually perform well with their defaults. I am not worried about setting parameters, I just want to try out tinymodels.
library(ranger)
glm_spec <- logistic_reg() %>%
set_engine('glm')
rf_spec <- rand_forest() %>%
set_mode('classification') %>%
set_engine('ranger')Now it’s time to get this into tinymodels and all working together. I am going to use workflow for this. workflow is a pre-processor and a model.
penguin_wf <- workflow() %>%
add_formula(sex ~ .)
penguin_wf══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Formula
Model: None
── Preprocessor ────────────────────────────────────────────────────────────────
sex ~ .So notice that when we call this we have a pre=processor, and no model. There is no model yet because I am going to use this to call two times to build both models. I can reuse this workflow as many times as I want for different models which is neat!
glm_results <- penguin_wf %>%
add_model(glm_spec) %>%
# fit(data = penguins_df) I could do this which would fit the model one time, but I want to use all those resamples I made so I will use fit_resamples instead
fit_resamples(resamples = penguin_boot,
control = control_resamples(save_pred = T, verbose = T)
)
glm_results# Resampling results
# Bootstrap sampling
# A tibble: 25 × 5
splits id .metrics .notes .predictions
<list> <chr> <list> <list> <list>
1 <split [249/91]> Bootstrap01 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
2 <split [249/89]> Bootstrap02 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
3 <split [249/89]> Bootstrap03 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
4 <split [249/84]> Bootstrap04 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
5 <split [249/93]> Bootstrap05 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
6 <split [249/82]> Bootstrap06 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
7 <split [249/89]> Bootstrap07 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
8 <split [249/94]> Bootstrap08 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
9 <split [249/92]> Bootstrap09 <tibble [2 × 4]> <tibble [1 × 3]> <tibble>
10 <split [249/96]> Bootstrap10 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
# ℹ 15 more rows
There were issues with some computations:
- Warning(s) x2: glm.fit: fitted probabilities numerically 0 or 1 occurred
Run `show_notes(.Last.tune.result)` for more information.rf_results <- penguin_wf %>%
add_model(rf_spec) %>%
fit_resamples(resamples = penguin_boot,
control = control_resamples(save_pred = T, verbose = T)
)
rf_results# Resampling results
# Bootstrap sampling
# A tibble: 25 × 5
splits id .metrics .notes .predictions
<list> <chr> <list> <list> <list>
1 <split [249/91]> Bootstrap01 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
2 <split [249/89]> Bootstrap02 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
3 <split [249/89]> Bootstrap03 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
4 <split [249/84]> Bootstrap04 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
5 <split [249/93]> Bootstrap05 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
6 <split [249/82]> Bootstrap06 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
7 <split [249/89]> Bootstrap07 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
8 <split [249/94]> Bootstrap08 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
9 <split [249/92]> Bootstrap09 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
10 <split [249/96]> Bootstrap10 <tibble [2 × 4]> <tibble [0 × 3]> <tibble>
# ℹ 15 more rowsNow to compare how they did.
collect_metrics(rf_results)# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.901 25 0.00426 Preprocessor1_Model1
2 roc_auc binary 0.964 25 0.00300 Preprocessor1_Model1# vs
collect_metrics(glm_results)# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.910 25 0.00522 Preprocessor1_Model1
2 roc_auc binary 0.975 25 0.00239 Preprocessor1_Model1These are very close together, perhaps the same. In this case I’d pick the linear model as it’s easier to interpret and faster to deploy. Since I saved the predictions we can start to look at them in more depth for the linear model.
glm_results %>%
conf_mat_resampled()# A tibble: 4 × 3
Prediction Truth Freq
<fct> <fct> <dbl>
1 female female 40.7
2 female male 4.2
3 male female 4.08
4 male male 42.2 glm_results %>%
collect_predictions() %>%
group_by(id) %>%
roc_curve(sex, .pred_female) %>%
ggplot(aes(1 - specificity, sensitivity, color = id)) +
geom_abline(lty = 2, color = 'gray80', size = 1.5) +
geom_path(show.legend = F, alpha = 0.9) +
coord_equal()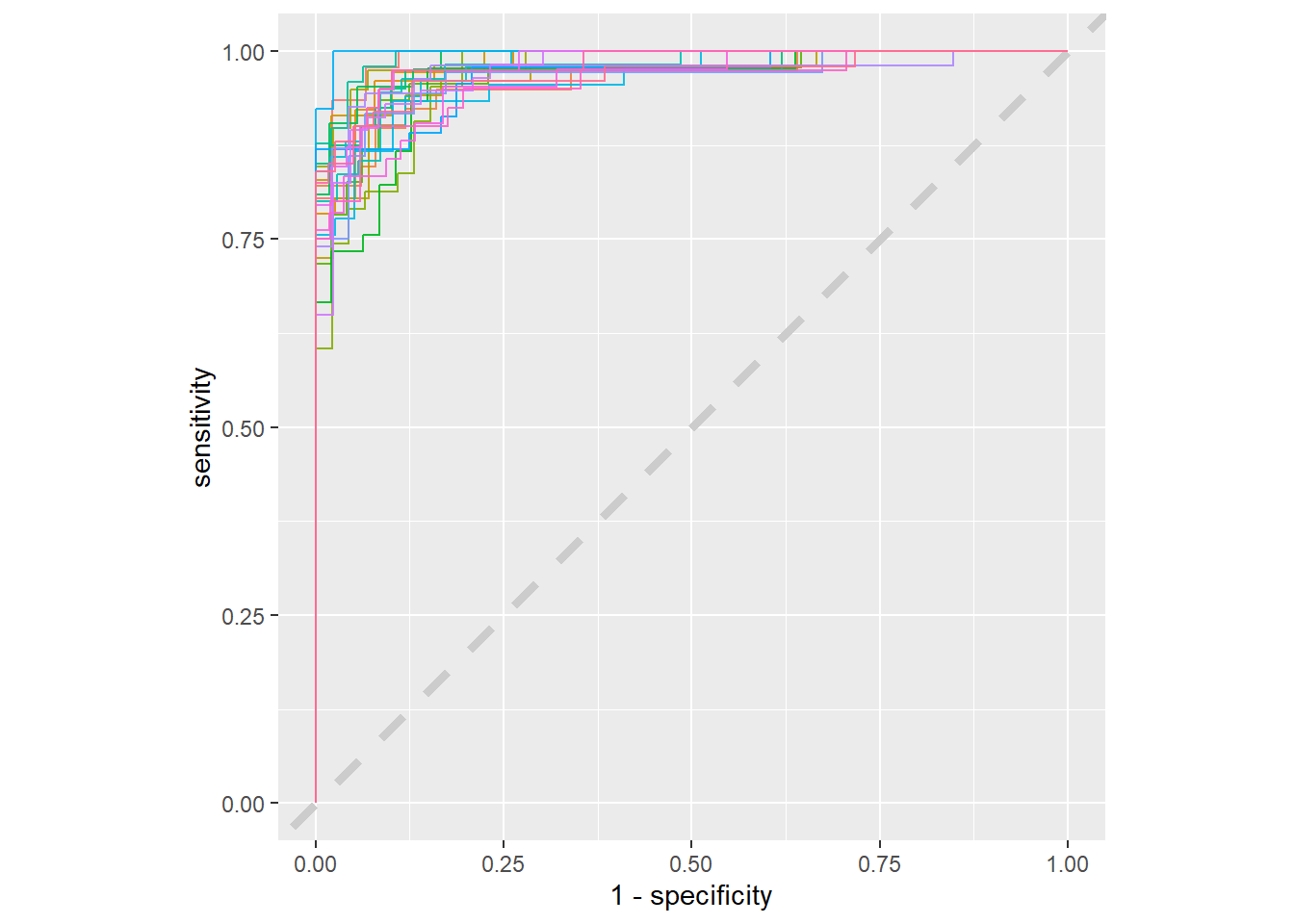
Having more data would make this less jagged, but this gives us an understanding of what the variation is from resample to resample. Things look pretty good, everything is fairly clustered. It passes the smell test. Now it’s finally time to come back to the testing data to estimate the performance on new data. I’ll use the last_fit function to fit the last and best fit to the testing data.
penguin_final <- penguin_wf %>%
add_model(glm_spec) %>%
last_fit(penguin_split)
collect_metrics(penguin_final)# A tibble: 2 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 accuracy binary 0.893 Preprocessor1_Model1
2 roc_auc binary 0.967 Preprocessor1_Model1collect_predictions(penguin_final) %>%
conf_mat(sex, .pred_class) Truth
Prediction female male
female 40 7
male 2 35Not to bad, I can see why some of the females would be classified as males as I did note the overlap in the exploration section. Overall this took less than an hour using packages I’ve never used and having to read the documentation for the workflow of these packages.
I want to take a look at a fitted workflow to see what appears in there now compared to when I last looked at a workflow, and we can look at the odds ratios to see how to interpret this model.
penguin_final$.workflow[[1]]══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Formula
Model: logistic_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
sex ~ .
── Model ───────────────────────────────────────────────────────────────────────
Call: stats::glm(formula = ..y ~ ., family = stats::binomial, data = data)
Coefficients:
(Intercept) speciesChinstrap speciesGentoo bill_length_mm
-91.797559 -8.434696 -11.983135 0.616936
bill_depth_mm flipper_length_mm body_mass_g
1.462226 0.085003 0.006811
Degrees of Freedom: 248 Total (i.e. Null); 242 Residual
Null Deviance: 345.2
Residual Deviance: 90.2 AIC: 104.2penguin_final$.workflow[[1]] %>%
tidy(exponentiate = T) %>%
arrange(estimate)# A tibble: 7 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 1.36e-40 16.0 -5.75 0.00000000900
2 speciesGentoo 6.25e- 6 3.27 -3.66 0.000253
3 speciesChinstrap 2.17e- 4 2.05 -4.12 0.0000373
4 body_mass_g 1.01e+ 0 0.00139 4.89 0.00000101
5 flipper_length_mm 1.09e+ 0 0.0546 1.56 0.120
6 bill_length_mm 1.85e+ 0 0.153 4.02 0.0000578
7 bill_depth_mm 4.32e+ 0 0.375 3.90 0.0000948 So for every 1 mm increase in bill depth for a penguin the odds of it being a male goes up over 4 times. So the bill depth is very important for penguins being male. I didn’t know that just bill depth would be such a good predictor. Let’s look at those plots from earlier now that we have the odds ratios.
penguins %>%
filter(!is.na(sex)) %>%
ggplot(aes(flipper_length_mm, bill_length_mm, color = sex, size = body_mass_g)) +
geom_point(alpha = .7) +
facet_wrap(~species)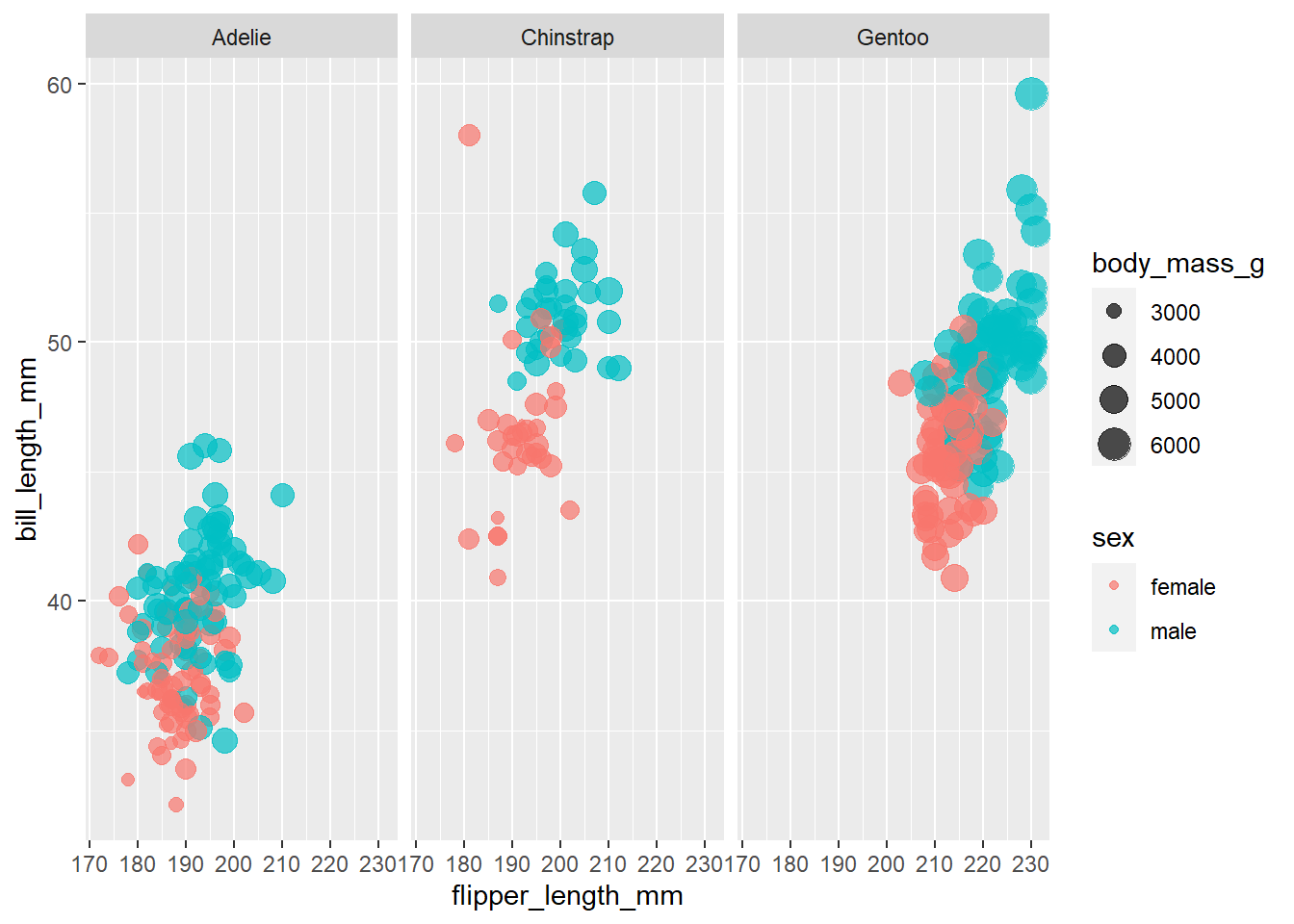
These are pretty overllaped, so now that we know the odds are much better for bill_depth_mm let’s put that on the x-axis and see if the data becomes less clustered.
penguins %>%
filter(!is.na(sex)) %>%
ggplot(aes(bill_depth_mm, bill_length_mm, color = sex, size = body_mass_g)) +
geom_point(alpha = .7) +
facet_wrap(~species)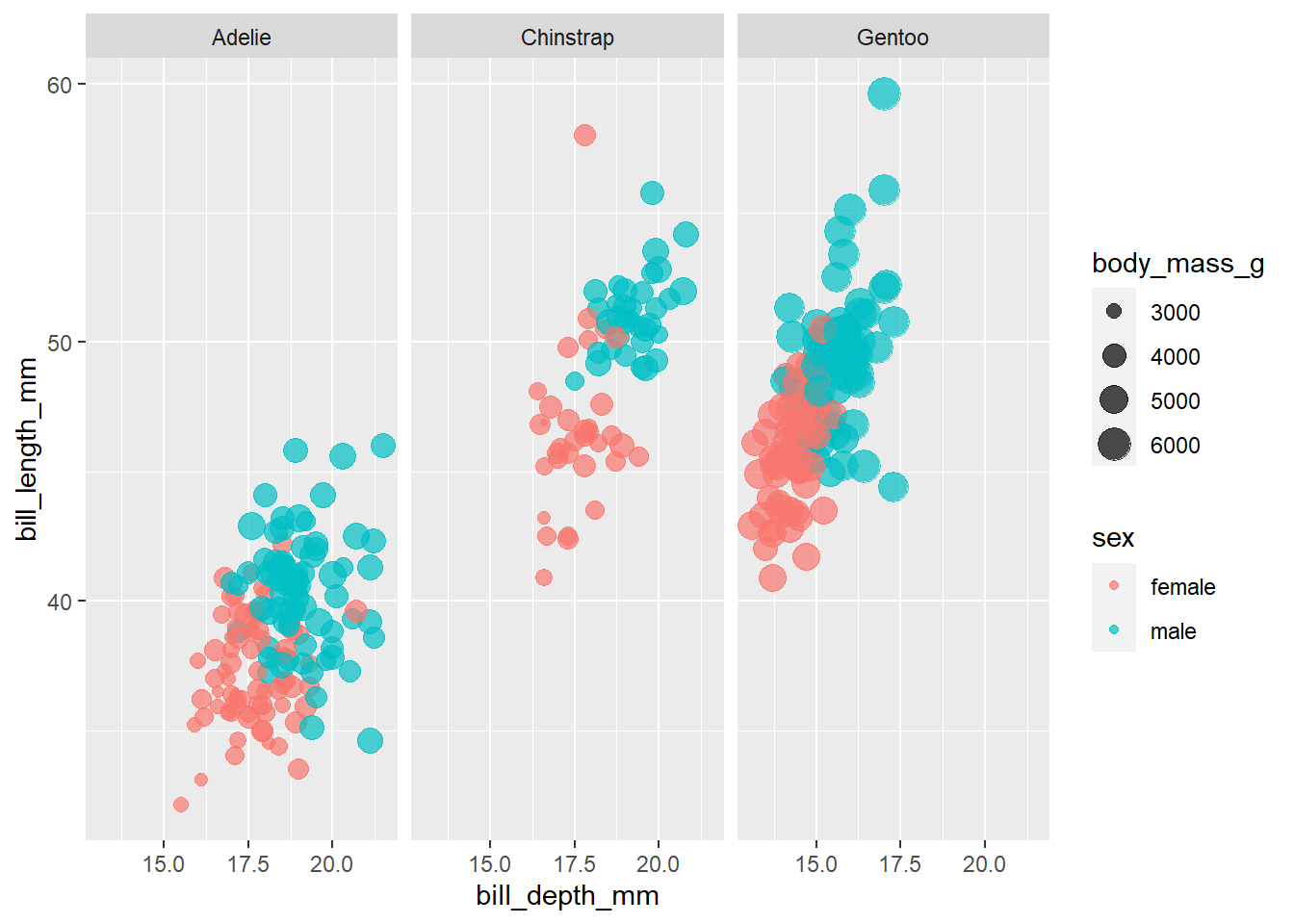
This does seem to seperate them out more dramatically than flipper_length_mm. Very cool.